You’ve decided to move on from the academic career path after finishing your masters or PhD. Congratulations! However, making the transition out of academia can be hard, intimidating, and lonely. There are so many possible paths, rather than the linear grad school to postdoc to faculty pipeline, and it can feel like you’re leaving your community behind after years in the university system. Here’s some advice that helped me with the transition to my first biotechnology job, and a few things I learned hiring scientists and managing a team at Loyal and Formic Labs. This advice is based on my own experience and the experiences of the people close to me – it won’t be perfectly applicable to fields outside of biotechnology. I’ll cover three key areas: how to find the right position, how to apply and get the job, and how to find your people.
How to find the right position
Narrow down your search space as much as possible
There are over three thousand biotech companies in the Bay Area alone. That’s a huge number compared to the 5-10 schools offering graduate biology degrees. Your first task is to narrow the search space using a few key factors.
- What field do you want to work in? Maybe your PhD research was in gene therapy delivery, and you’d like to stay in that space. Congrats, you just narrowed your search space down to only 88 companies in CA (data from BioPharmGuy, considering gene therapy, RNA and peptide therapy companies).
- What company size would you enjoy most? This can be a hard question to answer if you haven’t had a non-academic job before, but you can use clues from grad school. Knowing what you know now, what type of lab would you ideally want to work in? One with a small team and hands-on advisor, or a large lab with many graduate students and postdocs, but limited attention from your advisor? Are you excited or frightened by the idea of working in a new lab with a young advisor, before they’ve gotten tenure? The answers to these questions can steer you towards small and big companies, and towards or away from startups.
- Where do you want to live? Geography is an important consideration that shouldn’t be ignored. You now have the flexibility of being independent of the university system – use it to make a choice based on cost of living, proximity to family and friends, hobbies, or the best place to raise a family. Depending on the industry, your best options may be in one of a few hubs.
- Do you want to work remotely? If you enjoy the tradeoffs of remote work, limit your search to positions that offer this up front. Companies will often bring the entire team together a few times a year, so be prepared to travel at least at least a few times if you go down this route.
Talk to as many people as you can
You can start this process while you’re still in grad school. It’s not uncommon or uncool to do “informational interviews” with people in your field. These people might be a lab or university alumni, someone who has published in the same research area, or even just someone you follow online. I’ve had great luck in reaching out to strangers on Twitter or Linkedin to talk about ideas and careers.
Search smarter, not harder
Two websites I’ve already linked hold databases of biotech companies and a biotech-specific job board: BioPharmGuy and BioSpace. Searching on these sites can be great for both company discovery and job postings. AngelList Talent can help with the search for jobs at newer startups.
Get on Twitter
Twitter is a hub for science information, new publications, job postings, and gossip in the field. Especially for the startup scene, Twitter has far more value than Linkedin. You don’t even have to post anything, just find some interesting people to follow and go from there. The #AltAcChats hashtag is a good place to start.
Your skills are general – it’s okay to change fields
The skills you learn during a PhD are more generally applicable than you may believe. Did you manage projects involving several lab members or outside collaborators? Did you mentor undergrads or new members of the lab? TA and develop material for a course? Take on a project in a new research area after jumping into the deep end of the literature pool? Recognize, promote, and sell these skills – they are valuable in any field you end up committed to. Conquering a PhD means you can learn pretty much anything.
Get connected with the venture capitalists
The best VCs have an expert birds-eye-view of their industry, and they have an incentive to place talented people at their portfolio companies. I’ve talked with VCs from Lux Capital, 8VC, Northpond and others at biotech meetups. They’re always looking to network with talented people – they need dealflow just as much as you need a job or a term sheet!
Consider roles outside of pure research
Consider strategic operations, chief of staff, project management, VC, and other “alternative” roles. If you love being involved with science but don’t see yourself doing pure research forever, there are many ways to stay involved without opening a lab notebook.
How to apply and get the job
Your resume, cover letter, or intro needs to stand out
If you’ve identified a company and role that is a good fit for you, and you want to apply, realize that hiring managers get A LOT of resumes. This is especially true when a job is posted on Linkedin or other general job sites. If a manager only has a minute or two to devote to each resume, you have to stand out in a positive way. Maybe it’s a relevant and interesting thesis title, an open source software project you’ve contributed to, or a good word from someone working at the company. Any positive connection or good word can go a long way to getting you a first interview.
Do many, many interviews
Especially if you’re unfamiliar with the interview process, or they make you nervous. It might seriously suck at first, but the only way to get more comfortable with interviewing is to put yourself out there and get uncomfortable. In the age of zoom, you can interview with a company halfway across the country without ever leaving your room (or putting on pants). I’ll even suggest doing earlier interviews with companies that you may be a good fit for, but you know you wouldn’t take. You’ll learn some of the common interview questions, get practice summarizing your research experience, and learn about the salary bands for the role (you are going to ask about salary, right?)
Have something to show in public, especially if you’re interviewing for a computational or software role
This could be a personal website, Github repository, a website for a side project, or a reproducible demo analysis from a paper. You want something that can show off your programming and quantitative skills from any device connected to the internet. Be prepared to walk through design choices for the code and any areas that were particularly interesting or challenging. Good documentation is important for any software intended to be re-used – docs are valued more in industry than in academia.
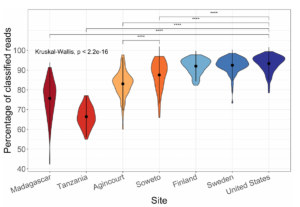
I have a few personal examples that I’ve repeatedly sent in messages or brought up live on a zoom interview. The bhattlab_workflows and kraken2_classification pipelines are not miracles of software engineering by any means, but they’re still used by members of the Bhatt lab and others, they make nice figures, and they have good docs. My bioinformatics in the cloud post is now a few years out of date, but it shows that I have been thinking about the challenges and solutions in this field for a while.
Brush up on the latest trends, languages, and frameworks in your field
In bioinformatics, Nextflow is the most popular workflow manager, and cloud compute skills are a necessity. Being familiar with both of these tools will help any bioinformatics interview. So, re-write a simple pipeline from grad school in Nextflow, sign up for the AWS Free Tier, and learn how to deploy it to AWS Batch. You could even write a blog post or a Twitter thread about the process, what you learned, and what you found challenging, then refer to it during an interview. A weekend of work will set you apart from those who haven’t tried to make the transition.
Utilize resources at your university
Many universities have free career counseling or job boards for people in situations just like you! Make sure you take advantage of these resources. You could probably benefit from a resume review, Linkedin profile checkup, or just someone knowledgeable to talk through your different options with.
Know what you’re worth. Negotiate.
Salary and equity compensation is field and role dependent. Talking with others in positions you’re applying to is the best way to get the current numbers. Ask for a range rather than direct numbers to avoid getting too personal. Also, recognize the tradeoffs that come with company size. Startups can’t pay as well, but can compensate with equity that could be life-changing in the event of a successful exit. Later-stage or public companies will be more stable and offer more in salary without the asymmetric upside. Finally, realize an offer is just a starting point for negotiations. There’s a hard limit for every position, but most offers can be flexed for the right candidate. You can also trade salary for equity (and vice-versa) depending on your risk tolerance.
How to find your people
Find your in-person community
There are growing meetup groups for young scientists in biotech and other fields. Right now, I’m seeing these mostly advertised in the Bay Area, NYC, and Boston, but they’re rapidly expanding to other areas as well. My top two for the Bay are Bits in Bio (which also has an active Slack community with over 2000 members) and Ergo Bio’s Biotech Venture Meetups. Groups like Nucleate bring together biotech founders from around the world.
Find your online community
I feel like the network of people talking about industry jobs, trends, and advice is stronger than ever. Twitter and Slack spaces like Bits in Bio are full of friendly and talented people.
Don’t stress about finding the “perfect” industry position in your first role out of grad school
Industry is not like academia, where you must commit 4+ years to a single field, and where your life is defined by your research area. You will learn more than you expect in the first year of your new role, and if you’re not happy, you’ll be in a better place to change it a year in. It’s much easier to change jobs in industry, and each change can come with better fit and increased compensation.