In the Neretti Lab @ Brown we are interested in the genomic and chromosome architecture changes that accompany replicative senescence. We studied this irreversible growth arrest in human diploid fibroblast (HDF) cells through the high-throughput chromatin conformation capture method Hi-C.
We compared Hi-C interaction data from healthy proliferating, synchronized quiescent and fully senescent cells. The main takeaways from the experiment were as follows.
- A general decrease in long-range chromatin contacts in senescence. We found that chromosomes in senescent cells are more likely to participate in short-range contacts when compared with proliferating. This mens that stable, long-range interactions are less likely and that the chromatin has become more compact. Importantly, we validated this result with several independent biochemical assays, including 3D-FISH, FAIRE and chromosome painting.
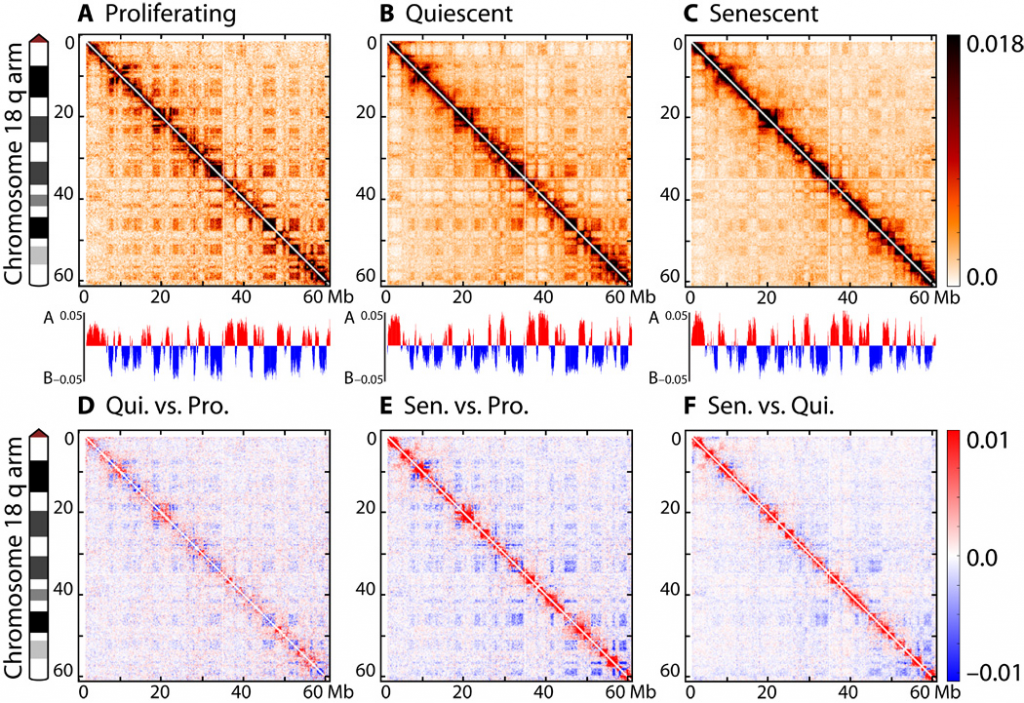
Top: Hi-C interaction matrices for proliferating, quiescent and senescent cells. Darker colors along the diagonal, and lighter colors off the diagonal in senescent indicate a loss of long-range interactions. Bottom: Subtracted interaction matrices show more interactions near the diagonal (short-range) in senescent cells.
- Chromatin domains that switch from an active compartment to a inactive compartment, or vice-versa. A subset of topologically associated domains, TADs, switch A/B compartments in senescence. This is interesting because the cells could theoretically use this method to direct transcription of senescence-associated genes, something we found by comparing with previously annotated gene sets.
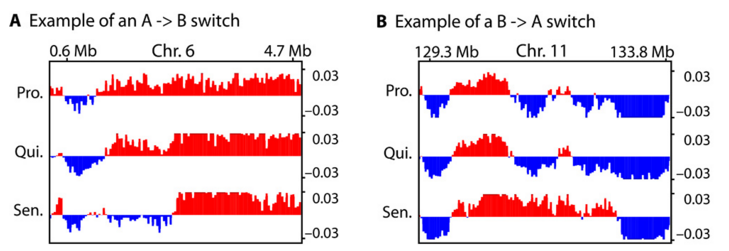
An example of TADs that switch compartments in senescence. Look at the transition of the first eigenvector signal from red to blue, or blue to red.
Our paper detailing this research was recently published in Science Advances [1]. Check it out here! I did a lot of computational work for this research, taking the data from raw sequences to normalized Hi-C contact maps, and finally interpreting them in various ways, so it feels great to have the results published. We were also featured in Brown’s science news.
[1] Criscione, S. W., De Cecco, M., Siranosian, B., Zhang, Y., Kreiling, J. A., Sedivy, J. M., & Neretti, N. (2016). Reorganization of chromosome architecture in replicative cellular senescence. Science Advances, 2(2), e1500882.