Update! This work has been published in Nature Communications.
Siranosian, B.A., Tamburini, F.B., Sherlock, G. et al. Acquisition, transmission and strain diversity of human gut-colonizing crAss-like phages. Nat Commun 11, 280 (2020). https://doi.org/10.1038/s41467-019-14103-3
Big questions in the microbiome field surround the topic of microbiome acquisition. Where do we get our first microbes from? What determines the microbes that colonize our guts form birth, and how do they change over time? What short and long term impacts do these microbes have on the immune system, allergies or diseases? What impact do delivery mode and breastfeeding have on the infant microbiome?
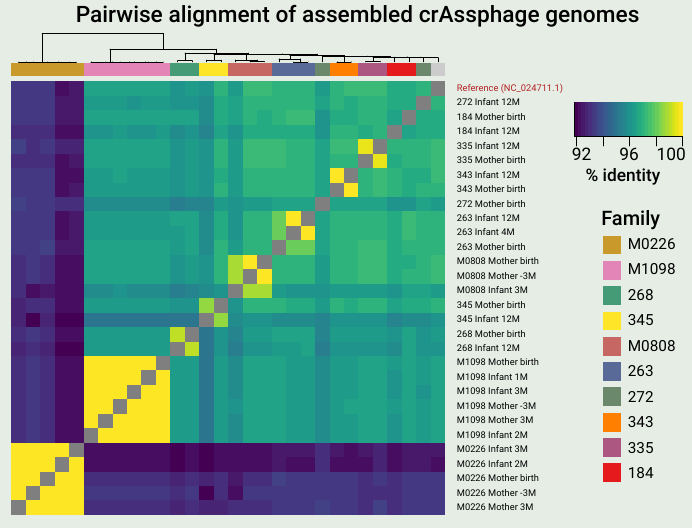
A key finding from the work was that mothers and infants often share identical or nearly identical crAssphage sequences, suggesting direct vertical transmission. Also, I love heatmaps.
As you might expect, a major source for microbes colonizing the infant gut is immediate family members, and the mother is thought to be the major source. Thanks to foundational studies by Bäckhed, Feretti, Yassour and others (references below), we now know that infants often acquire the primary bacterial strain from the mother’s microbiome. These microbes can have beneficial capabilities for the infant, such as the ability to digest human milk oligosaccharides, a key source of nutrients in breast milk.
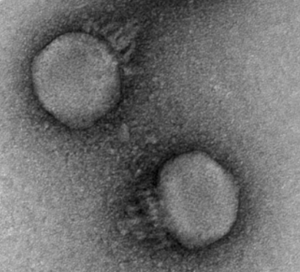
The microbiome isn’t just bacteria – phages (along with fungi and archaea to a smaller extent) play key roles. Phages are viruses that predate on bacteria, depleting certain populations and exchanging genes among the bacteria they infect. Interestingly, phages were previously shown to display different inheritance patterns than bacteria, remaining individual-specific between family members and even twins (Reyes et al. 2010). CrAss-like phages are the most prevalent and abundant group of phages colonizing the human gut, and our lab was interested in the inheritance patterns of these phages.
We examined publicly available shotgun gut metagenomic datasets from two studies (Yassour et al. 2018, Bäckhed et al. 2015), containing 134 mother-infant pairs sampled extensively through the first year of life. In contrast to what has been observed for other members of the gut virome, we observed many putative transmission events of a crAss-like phage from mother to infant. The key takeaways from our research are summarized below. You can also refer my poster from the Cold Spring Harbor Microbiome meeting for the figures supporting these points. We hope to have a new preprint (and hopefully a publication) on this research out soon!
- CrAssphage is not detected in infant microbiomes at birth, increases in prevalence with age, but doesn’t reach the level of adults by 12 months of age.
- Mothers and infants share nearly identical crAssphage genomes in 40% of cases, suggesting vertical transmission.
- Infants have reduced crAssphage strain diversity and typically acquire the mother’s dominant strain upon transmission.
- Strain diversity is mostly the result of neutral genetic variation, but infants have more nonsynonymous multiallelic sites than mothers.
- Strain diversity varies across the genome, and tail fiber genes are enriched for strain diversity with nonsynonymous variants.
- These findings extend to crAss-like phages. Vaginally born infants are more likely to have crAss-lke phages than those born via C-section.
References
1. Reyes, A. et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010).
2. Yassour, M. et al. Strain-Level Analysis of Mother-to-Child Bacterial Transmission during the First Few Months of Life. Cell Host & Microbe 24, 146-154.e4 (2018).
3. Bäckhed, F. et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host & Microbe 17, 690–703 (2015).
4. Ferretti, P. et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host & Microbe 24, 133-145.e5 (2018).