CrAssphage is a like mystery novel full of surprises. First described in 2014 by Dutilh et al., crAssphage acquired it’s (rather unfortunate, given that it colonizes the human intestine) name from the “Cross-Assembly” bioinformatics method used to characterize it. CrAssphage interests me because it’s prevalent in up to 70% of human gut microbiomes, and can make up the majority of viral sequencing reads in a metagenomics experiment. This makes it the most successful single entity colonizing human microbiomes. However, no health impacts have been demonstrated from having crAssphage in your gut – several studies (Edwards et al. 2019) have turned up negative.
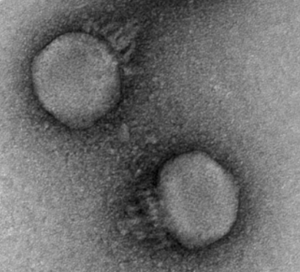
Electron micrograph of a representative crAssphage, from Shkoporov et al. (2018). This phage is a member of the Podoviridae family and infects Bacteroides Intestinalis.
CrAssphage was always suspected to predate on species of the Bacteroides genus based on evidence from abundance correlation and CRISPR spacers. However, the phage proved difficult to isolate and culture. It wasn’t until recently that a crAssphage was confirmed to infect Bacteroides intestinalis (Shakoporov et al. 2018). They also got a great TEM image of the phage! With crAssphage successfully cultured in the lab, scientists have begun to answer fundamental questions about its biology. The phage appears to have a narrow host range, infecting a single B. intestinalis strain and not others or other species. The life cycle of the phage was puzzling:
“We can conclude that the virus probably causes a successful lytic infection with a size of progeny per capita higher than 2.5 in a subset of infected cells (giving rise to a false overall burst size of ~2.5), and also enters an alternative interaction (pseudolysogeny, dormant, carrier state, etc.) with some or all of the remaining cells. Overall, this allows both bacteriophage and host to co-exist in a stable interaction over prolonged passages. The nature of this interaction warrants further investigation.” (Shakoporov et al. 2018)
Further investigation showed that crAssphage is one member of an extensive family of “crAss-like” phages colonizing the human gut. Guerin et al. (2018) proposed a classification system for these phages, which contains 4 subfamilies (Alpha, Beta, Delta and Gamma) and 10 clusters. The first described “prototypical crAssphage” belongs to the Alpha subfamily, cluster 1. It struck me how diverse these phages are – different families are less than 20% identical at the protein level! When all crAss-like phages are considered, it’s estimated that up to 100% of individuals cary at least one crAss-like phage, and most people cary more than one.
Given the high prevalence of crAss-like phages and their specificity for the human gut, they do have an interesting use as a tracking device for human sewage. DNA from crAss-like phages can be used to track waste contamination into water, for example (Stachler et al. 2018). In a similar vein, our lab has used crAss-like phages to better understand how microbes are transmitted from mothers to newborn infants. The small genome sizes (around 100kb) and high prevalence/abundance make these phages good tools for doing strain-resolved metagenomics. Trust me, you’d much rather do genomic assembly and variant calling on a 100kb phage genome than a 3Mb bacterial genome!
Research into crAss-like phages is just beginning, and I’m excited to see what’s uncovered in the future. What are the hosts of the various phage clusters? How do these phages influence gut bacterial communities? Do crAss-like phages exclude other closely related phages from colonizing their niches, leading to the low strain diversity we observe? Can crAss-like phgaes be used to engineer bacteria in the microbiome, delivering precise genetic payloads? This final question in the most interesting to me, given that crAss-like phages seem relatively benign to humans, yet incredibly capable of infecting our microbes.
References
1.Dutilh, B. E. et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nature Communications 5, 4498 (2014).
2.Edwards, R. A. et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nature Microbiology 1 (2019). doi:10.1038/s41564-019-0494-6
3.Guerin, E. et al. Biology and Taxonomy of crAss-like Bacteriophages, the Most Abundant Virus in the Human Gut. Cell Host & Microbe 0, (2018).
4.Shkoporov, A. N. et al. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nature Communications 9, 4781 (2018).
5.Stachler, E., Akyon, B., de Carvalho, N. A., Ference, C. & Bibby, K. Correlation of crAssphage qPCR Markers with Culturable and Molecular Indicators of Human Fecal Pollution in an Impacted Urban Watershed. Environ. Sci. Technol. 52, 7505–7512 (2018).